AutoCloner is a custom pipeline that designs homeologue-specific primers for polyploids. It achieves this by extracting SNP locations unique to your gene of interest, and placing the 3` ends of primers at these SNPs.
Whilst there is an existing tool for designing KASP primers in wheat, PolyMarker (Ramirez-Gonzalez et al., 2015), there is not currently anything available for researchers who wish to clone entire genes. This is where AutoCloner comes in.
Please input your sequence in the form to the right. AutoCloner will design a stacked PCR arrangement of primers depending on your specified minimum and maximum product lengths.
Source code and documentation at GitHub.
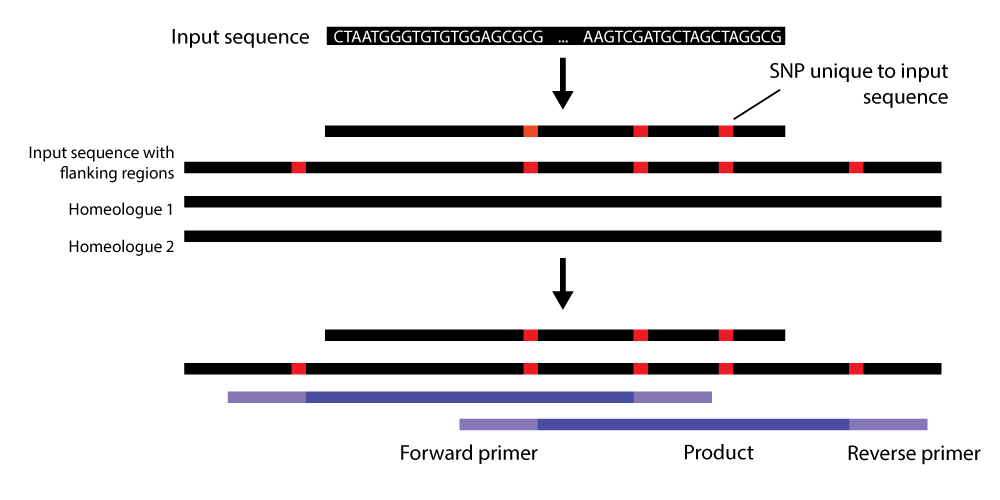
Fig. 1 Illustration of the AutoCloner pipeline. Flanking regions are extracted from the genome before mining for SNPs and subsequent primer design.

Based at the University of Bristol with support from BBSRC.


AutoCloner maintained by Alex Coulton.